Linear Models
Contents
Linear Models#
Overview#
This notebook contains an initial exploration of different linear regressions to predict monthly ED demand.
For all models, variables used include:
Service capacity (111, GP, Ambulance)
Service utility (111, Ambulance)
Population
Population Health (People, Places, Lives)
#turn warnings off to keep notebook tidy
import warnings
warnings.filterwarnings('ignore')
Import libraries#
import os
import pandas as pd
import numpy as np
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import Lasso
from sklearn.linear_model import LassoLarsIC
from sklearn.linear_model import Ridge
from sklearn.linear_model import ElasticNet
from sklearn.linear_model import Lars
from sklearn.linear_model import OrthogonalMatchingPursuit as OMP
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import train_test_split
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import cross_validate
from sklearn.model_selection import RepeatedKFold
from sklearn.model_selection import KFold
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import RFECV
from sklearn.feature_selection import RFE
from sklearn.feature_selection import mutual_info_regression
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline
plt.style.use('ggplot')
Import data#
dta = pd.read_csv('https://raw.githubusercontent.com/CharlotteJames/ed-forecast/main/data/master_scaled_new.csv',
index_col=0)
dta.columns = ['_'.join([c.split('/')[0],c.split('/')[-1]])
if '/' in c else c for c in dta.columns]
dta.ccg.unique().shape
(74,)
Add random feature#
# Adding random features
rng = np.random.RandomState(0)
rand_var = rng.rand(dta.shape[0])
dta['rand1'] = rand_var
dta.shape
(1618, 14)
Train test split#
train, test = train_test_split(dta,random_state=29)
y = dta['ae_attendances_attendances']
X = dta.drop(['ae_attendances_attendances','ccg','month'], axis=1)
y_train = train['ae_attendances_attendances'].values
X_train = train.drop(['ae_attendances_attendances','ccg','month'], axis=1)
y_test = test['ae_attendances_attendances'].values
X_test = test.drop(['ae_attendances_attendances','ccg','month'], axis=1)
Ridge regression#
No scaling#
model = Ridge()
#model = make_pipeline(StandardScaler(), RidgeCV())
model.fit(X_train, y_train)
print(f'model score on training data: {model.score(X_train, y_train)}')
print(f'model score on testing data: {model.score(X_test, y_test)}')
model score on training data: 0.5440194513191048
model score on testing data: 0.5336104269604207
coefs = pd.DataFrame(
model.coef_,
columns=['Coefficients'], index=X_train.columns
)
coefs.plot(kind='barh', figsize=(9, 7))
plt.title('Ridge model')
plt.axvline(x=0, color='.5')
plt.subplots_adjust(left=.3)
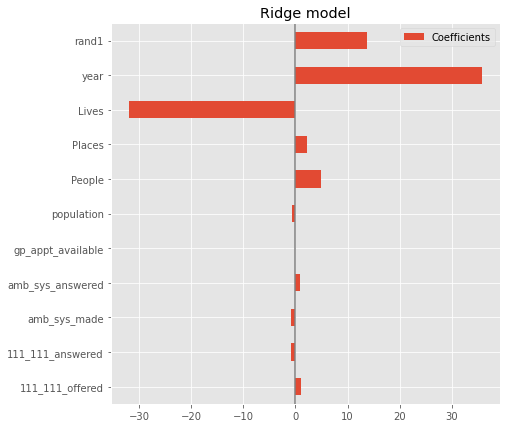
Without scaling features it is not possible to compare their importance using model coefficients
Standardise features#
model = make_pipeline(StandardScaler(), Ridge())
model.fit(X_train, y_train)
print(f'model score on training data: {model.score(X_train, y_train)}')
print(f'model score on testing data: {model.score(X_test, y_test)}')
model score on training data: 0.543812323234377
model score on testing data: 0.5336184653406383
coefs = pd.DataFrame(
model[1].coef_,
columns=['Coefficients'], index=X_train.columns
)
coefs.plot(kind='barh', figsize=(9, 7))
plt.title('Ridge model')
plt.axvline(x=0, color='.5')
plt.subplots_adjust(left=.3)
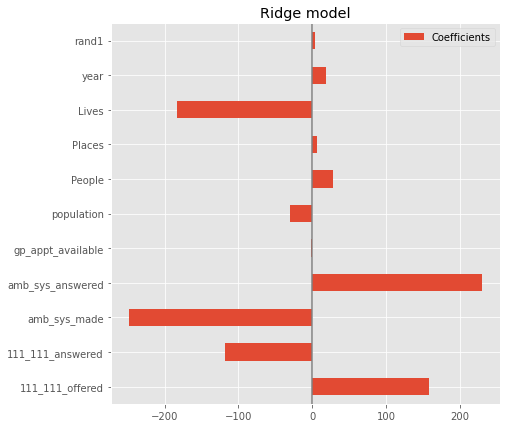
Feature selection#
selector = RFECV(model[1], step=1, cv=5)
selector = selector.fit(X_train, y_train)
support = pd.DataFrame(
selector.support_,
columns=['Support'], index=X_train.columns
)
support
| Support | |
|---|---|
| 111_111_offered | True |
| 111_111_answered | True |
| amb_sys_made | True |
| amb_sys_answered | True |
| gp_appt_available | False |
| population | True |
| People | True |
| Places | True |
| Lives | True |
| year | True |
| rand1 | True |
The random variable should not be supported
With cross-validation#
cv_model = cross_validate(
model, X,y,
cv=RepeatedKFold(n_splits=5, n_repeats=5, random_state=0),
return_estimator=True, n_jobs=2
)
coefs = pd.DataFrame(
[model[1].coef_
for model in cv_model['estimator']],
columns=X.columns
)
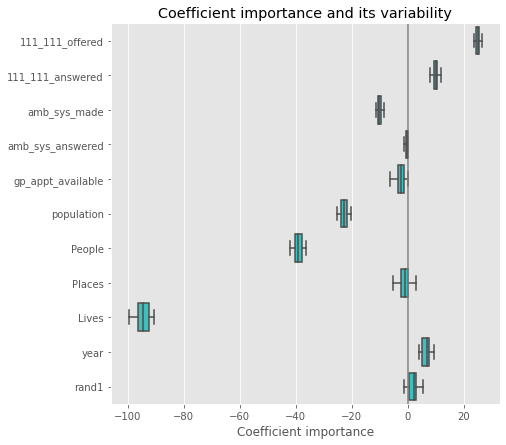
plt.figure(figsize=(9, 7))
sns.boxplot(data=coefs, orient='h', color='cyan', saturation=0.5)
plt.axvline(x=0, color='.5')
plt.xlabel('Coefficient importance')
plt.title('Coefficient importance and its variability')
plt.subplots_adjust(left=.3)
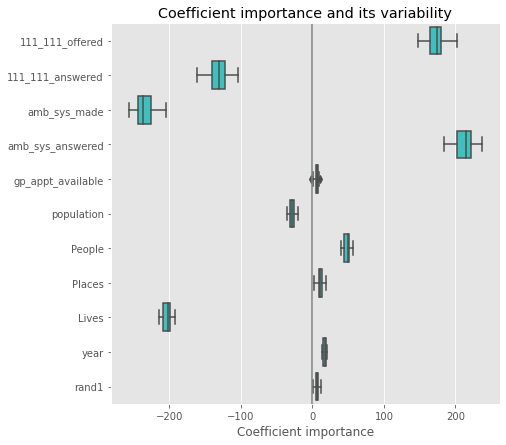
Lasso#
Standardise Features#
model = make_pipeline(StandardScaler(), Lasso(alpha=.015, max_iter=100000))
model.fit(X_train, y_train)
print(f'model score on training data: {model.score(X_train, y_train)}')
print(f'model score on testing data: {model.score(X_test, y_test)}')
model score on training data: 0.5440180818857432
model score on testing data: 0.5335852489275236
coefs = pd.DataFrame(
model[1].coef_,
columns=['Coefficients'], index=X_train.columns
)
coefs.plot(kind='barh', figsize=(9, 7))
plt.title('Lasso model, strong regularization')
plt.axvline(x=0, color='.5')
plt.subplots_adjust(left=.3)
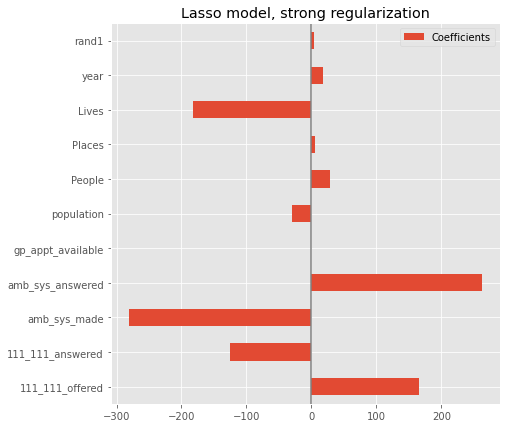
Feature selection#
selector = RFECV(model[1], step=1, cv=5)
selector = selector.fit(X_train, y_train)
support = pd.DataFrame(
selector.support_,
columns=['Support'], index=X_train.columns
)
support
| Support | |
|---|---|
| 111_111_offered | True |
| 111_111_answered | True |
| amb_sys_made | True |
| amb_sys_answered | True |
| gp_appt_available | False |
| population | True |
| People | True |
| Places | True |
| Lives | True |
| year | True |
| rand1 | True |
Cross validate#
cv_model = cross_validate(
model, X,y,
cv=RepeatedKFold(n_splits=5, n_repeats=5, random_state=0),
return_estimator=True, n_jobs=2
)
coefs = pd.DataFrame(
[model[1].coef_
for model in cv_model['estimator']],
columns=X.columns
)
plt.figure(figsize=(9, 7))
sns.boxplot(data=coefs, orient='h', color='cyan', saturation=0.5)
plt.axvline(x=0, color='.5')
plt.xlabel('Coefficient importance')
plt.title('Coefficient importance and its variability')
plt.subplots_adjust(left=.3)
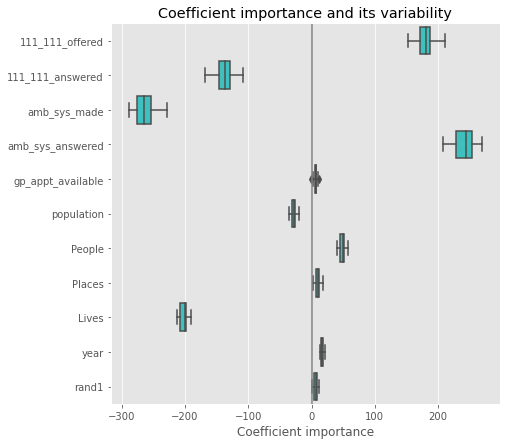
OLS#
Standardise Features#
model = make_pipeline(StandardScaler(), LinearRegression())
model.fit(X_train, y_train)
print(f'model score on training data: {model.score(X_train, y_train)}')
print(f'model score on testing data: {model.score(X_test, y_test)}')
model score on training data: 0.5440195223174749
model score on testing data: 0.5336193991596547
coefs = pd.DataFrame(
model[1].coef_,
columns=['Coefficients'], index=X_train.columns
)
coefs.plot(kind='barh', figsize=(9, 7))
plt.title('OLS')
plt.axvline(x=0, color='.5')
plt.subplots_adjust(left=.3)
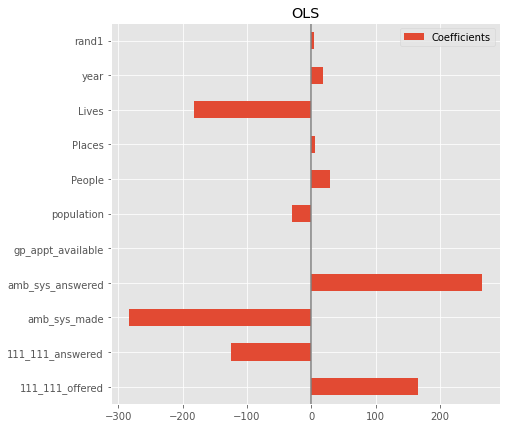
Feature selection#
selector = RFECV(model[1], step=1, cv=5)
selector = selector.fit(X_train, y_train)
support = pd.DataFrame(
selector.support_,
columns=['Support'], index=X_train.columns
)
support
| Support | |
|---|---|
| 111_111_offered | True |
| 111_111_answered | True |
| amb_sys_made | True |
| amb_sys_answered | True |
| gp_appt_available | False |
| population | True |
| People | True |
| Places | True |
| Lives | True |
| year | True |
| rand1 | True |
Cross validate#
cv_model = cross_validate(
model, X,y,
cv=RepeatedKFold(n_splits=5, n_repeats=5, random_state=0),
return_estimator=True, n_jobs=2
)
coefs = pd.DataFrame(
[model[1].coef_
for model in cv_model['estimator']],
columns=X.columns
)
plt.figure(figsize=(9, 7))
sns.boxplot(data=coefs, orient='h', color='cyan', saturation=0.5)
plt.axvline(x=0, color='.5')
plt.xlabel('Coefficient importance')
#plt.title('Coefficient importance and its variability')
plt.tight_layout()
plt.savefig('ols_feature_importance.pdf')
plt.subplots_adjust(left=.3)
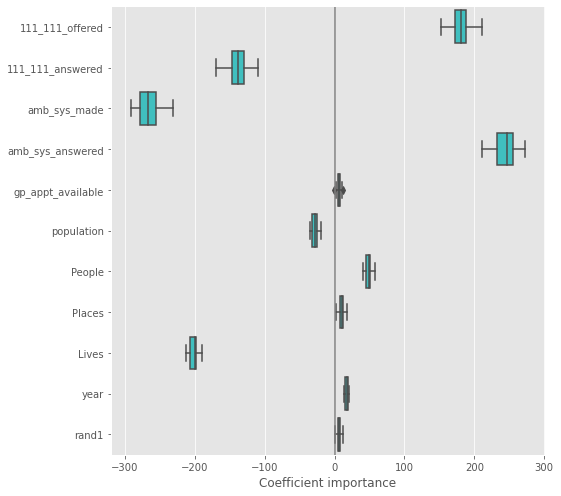
ElasticNet#
Standardise Features#
model = make_pipeline(StandardScaler(),
ElasticNet(max_iter=100000))
model.fit(X_train, y_train)
print(f'model score on training data: {model.score(X_train, y_train)}')
print(f'model score on testing data: {model.score(X_test, y_test)}')
model score on training data: 0.4727309935289348
model score on testing data: 0.43772822174451687
coefs = pd.DataFrame(
model[1].coef_,
columns=['Coefficients'], index=X_train.columns
)
coefs.plot(kind='barh', figsize=(9, 7))
plt.title('ElasticNet CV')
plt.axvline(x=0, color='.5')
plt.subplots_adjust(left=.3)
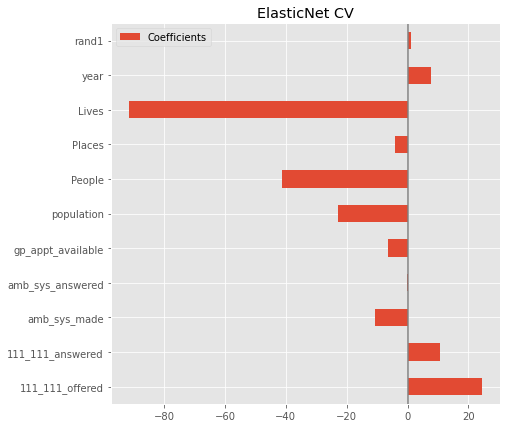
Cross validate#
cv_model = cross_validate(
model, X,y,
cv=RepeatedKFold(n_splits=5, n_repeats=5, random_state=0),
return_estimator=True, n_jobs=2
)
coefs = pd.DataFrame(
[model[1].coef_
for model in cv_model['estimator']],
columns=X.columns
)
plt.figure(figsize=(9, 7))
sns.boxplot(data=coefs, orient='h', color='cyan', saturation=0.5)
plt.axvline(x=0, color='.5')
plt.xlabel('Coefficient importance')
plt.title('Coefficient importance and its variability')
plt.subplots_adjust(left=.3)
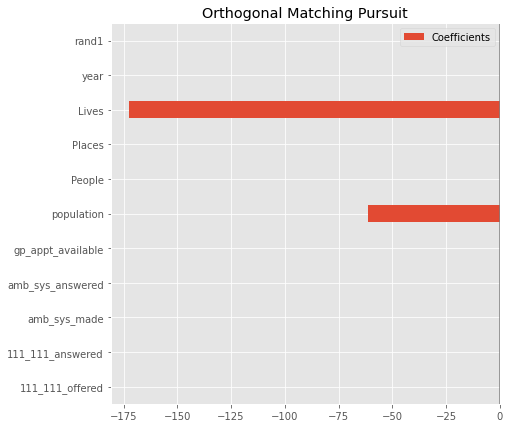
Feature Selection#
OMP#
model = make_pipeline(StandardScaler(),
OMP(n_nonzero_coefs = 2, normalize=False))
model.fit(X_train, y_train)
print(f'model score on training data: {model.score(X_train, y_train)}')
print(f'model score on testing data: {model.score(X_test, y_test)}')
model score on training data: 0.4992217993534437
model score on testing data: 0.4823928451338607
coefs = pd.DataFrame(
model[1].coef_,
columns=['Coefficients'], index=X_train.columns
)
coefs.plot(kind='barh', figsize=(9, 7))
plt.title('Orthogonal Matching Pursuit')
plt.axvline(x=0, color='.5')
plt.subplots_adjust(left=.3)
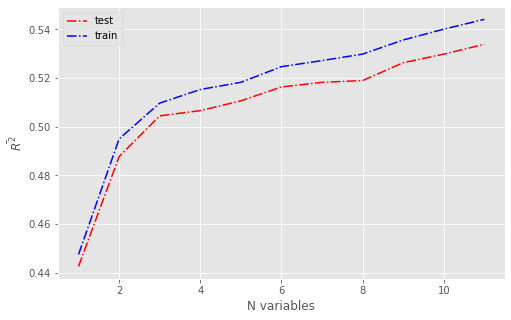
Score vs n_feat#
num_features = [i+1 for i in range(X.shape[1])]
results_train, results_test = pd.DataFrame(), pd.DataFrame()
for k in num_features:
model = make_pipeline(StandardScaler(),
OMP(n_nonzero_coefs = k, normalize=False))
cv = RepeatedKFold(n_splits=5, n_repeats=5, random_state=1)
scores_train, scores_test = [],[]
for train_index, test_index in cv.split(X, y):
model.fit(X.iloc[train_index], y.iloc[train_index])
scores_test.append(model.score(X.iloc[test_index], y.iloc[test_index]))
scores_train.append(model.score(X.iloc[train_index], y.iloc[train_index]))
results_test[k]=scores_test
results_train[k]=scores_train
fig = plt.figure(figsize=(8,5))
plt.plot(num_features, results_test.mean(), 'r-.', label='test')
plt.plot(num_features, results_train.mean(), 'b-.', label='train')
plt.xlabel('N variables')
plt.ylabel(r'$\bar{R^2}$')
plt.legend(loc='best')
#plt.savefig('rsq_features_omp.pdf')
plt.show()
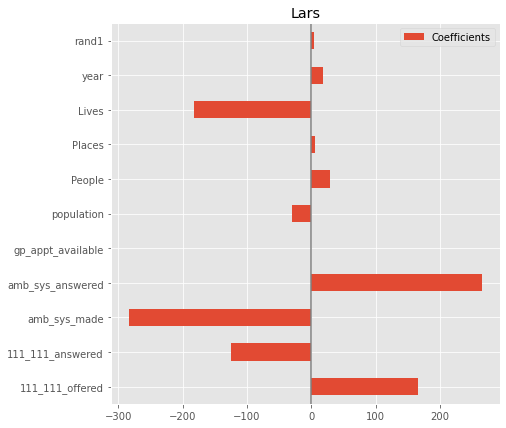
LARS#
from sklearn.linear_model import Lars
model = make_pipeline(StandardScaler(),
Lars(n_nonzero_coefs = X.shape[1], normalize=False))
model.fit(X_train, y_train)
print(f'model score on training data: {model.score(X_train, y_train)}')
print(f'model score on testing data: {model.score(X_test, y_test)}')
model score on training data: 0.5440195223174749
model score on testing data: 0.533619399159655
coefs = pd.DataFrame(
model[1].coef_,
columns=['Coefficients'], index=X_train.columns
)
coefs.plot(kind='barh', figsize=(9, 7))
plt.title('Lars')
plt.axvline(x=0, color='.5')
plt.subplots_adjust(left=.3)
Score vs number of features#
num_features = [i+1 for i in range(X.shape[1])]
results_train, results_test = pd.DataFrame(), pd.DataFrame()
for k in num_features:
model = make_pipeline(StandardScaler(),
Lars(n_nonzero_coefs = k, normalize=False))
cv = RepeatedKFold(n_splits=5, n_repeats=5, random_state=1)
scores_train, scores_test = [],[]
for train_index, test_index in cv.split(X, y):
model.fit(X.iloc[train_index], y.iloc[train_index])
scores_test.append(model.score(X.iloc[test_index], y.iloc[test_index]))
scores_train.append(model.score(X.iloc[train_index], y.iloc[train_index]))
results_test[k]=scores_test
results_train[k]=scores_train
fig = plt.figure(figsize=(8,5))
plt.plot(num_features, results_test.mean(), 'r-.', label='test')
plt.plot(num_features, results_train.mean(), 'b-.', label='train')
plt.xlabel('N variables')
plt.ylabel(r'$\bar{R^2}$')
plt.legend(loc='best')
#plt.savefig('rsq_features_lars.pdf')
plt.show()
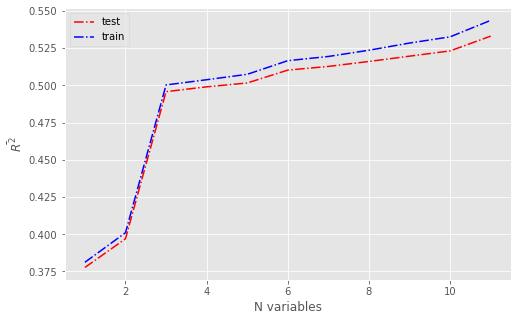
Mutual information#
mi = mutual_info_regression(X_train,y_train)
mi_df = pd.DataFrame(
mi,
columns=['Mutual Information'], index=X_train.columns
)
mi_df
| Mutual Information | |
|---|---|
| 111_111_offered | 0.368889 |
| 111_111_answered | 0.342691 |
| amb_sys_made | 0.618272 |
| amb_sys_answered | 0.716384 |
| gp_appt_available | 0.087838 |
| population | 1.394764 |
| People | 1.275716 |
| Places | 1.387013 |
| Lives | 1.317528 |
| year | 0.032590 |
| rand1 | 0.038655 |
Cross validated#
num_features = [i+1 for i in range(X.shape[1])]
results_train, results_test = pd.DataFrame(), pd.DataFrame()
for k in num_features:
fs = SelectKBest(score_func=mutual_info_regression, k=k)
model = make_pipeline(fs, LinearRegression())
cv = RepeatedKFold(n_splits=5, n_repeats=5, random_state=1)
scores_train, scores_test = [],[]
for train_index, test_index in cv.split(X, y):
model.fit(X.iloc[train_index], y.iloc[train_index])
scores_test.append(model.score(X.iloc[test_index], y.iloc[test_index]))
scores_train.append(model.score(X.iloc[train_index], y.iloc[train_index]))
results_test[k]=scores_test
results_train[k]=scores_train
print('>%d %.3f (%.3f)' % (k, np.mean(scores_test), np.std(scores_test)))
>1 0.077 (0.086)
>2 0.165 (0.066)
>3 0.496 (0.036)
>4 0.501 (0.035)
>5 0.502 (0.035)
>6 0.509 (0.035)
>7 0.524 (0.034)
>8 0.531 (0.033)
>9 0.531 (0.033)
>10 0.534 (0.034)
>11 0.534 (0.035)
fig,ax = plt.subplots(figsize=(8,5))
plt.plot(num_features, results_test.mean(), 'r-.', label='test')
plt.plot(num_features, results_train.mean(), 'b-.', label='train')
plt.fill_between(num_features, y1=(results_test.mean()-results_test.std()).values,
y2=(results_test.mean()+results_test.std()).values, alpha=0.2, color='r')
plt.fill_between(num_features, y1=results_train.mean()-results_train.std(),
y2=results_train.mean()+results_train.std(), alpha=0.2, color='b')
plt.xlabel('N variables')
plt.ylabel(r'$\bar{R^2}$')
plt.legend(loc='best')
#plt.savefig('rsq_features_mi.pdf')
plt.show()
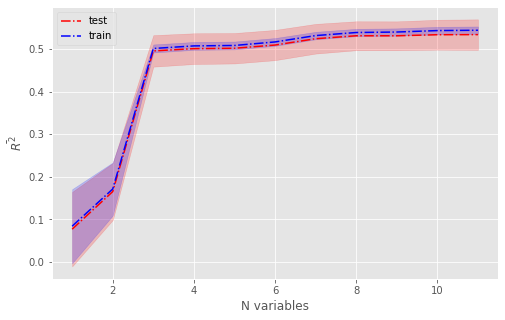
Average importance#
fs = SelectKBest(score_func=mutual_info_regression, k='all')
fs.fit(X,y)
mi_df = pd.DataFrame(
fs.scores_,
columns=['Scores'], index=X.columns
)
mi_df.sort_values(by='Scores', ascending=False)
| Scores | |
|---|---|
| Places | 1.436297 |
| population | 1.436055 |
| Lives | 1.391061 |
| People | 1.351514 |
| amb_sys_answered | 0.692218 |
| amb_sys_made | 0.628977 |
| 111_111_offered | 0.383233 |
| 111_111_answered | 0.338584 |
| gp_appt_available | 0.096549 |
| year | 0.021728 |
| rand1 | 0.007324 |
Summary#
Models perform comparatively well, with an \(R^2\) of ~ 0.56
The random feature is supported when using recursive feature elimination
When using the mutual information score, the random feature is excluded. Population and measures of population health are most important.
Mutual information accounts for non-linear relationships, suggesting a non-linear model may be more appropriate.

